publications
In Progress 🏗
2025
-
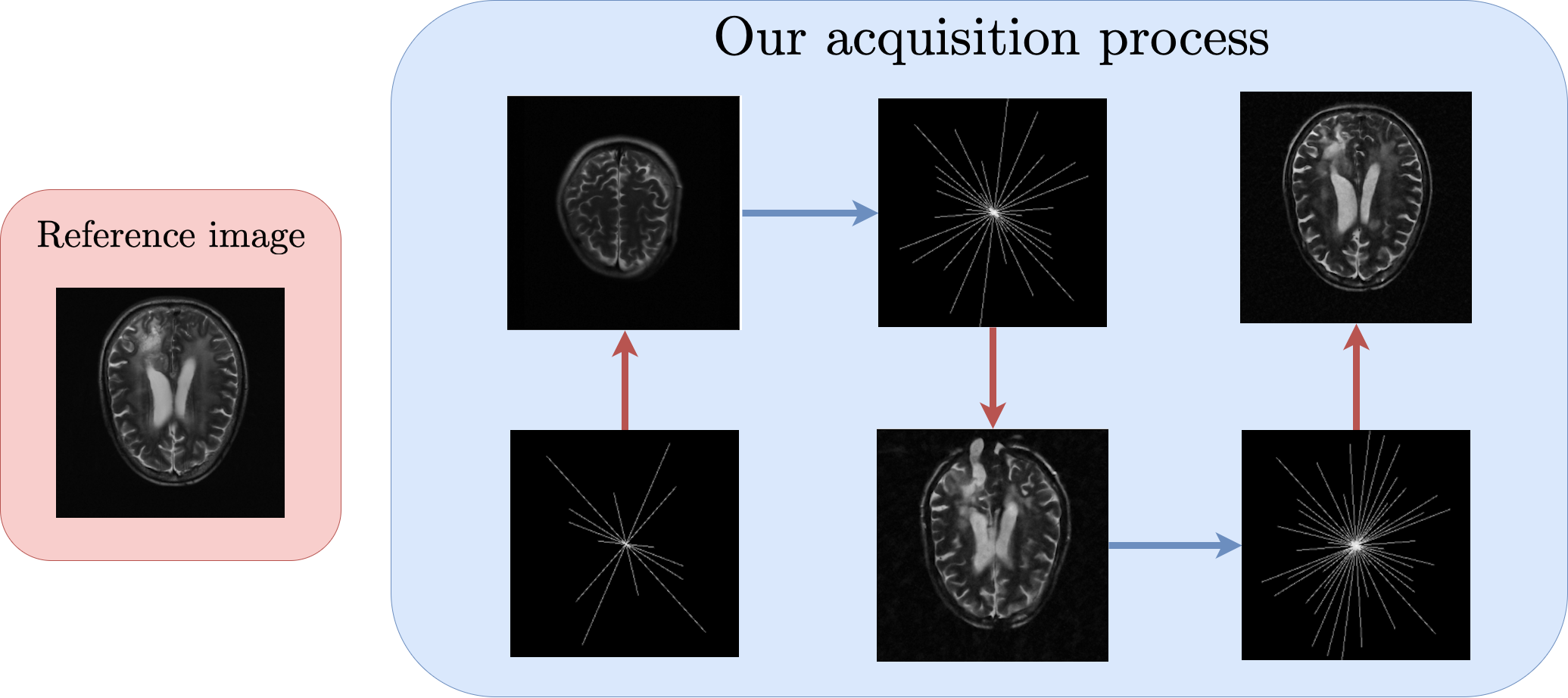 Active MRI Acquisition with Diffusion Guided Bayesian Experimental DesignJacopo Iollo, Geoffroy Oudoumanessah, Carole Lartizien, and 2 more authors2025
Active MRI Acquisition with Diffusion Guided Bayesian Experimental DesignJacopo Iollo, Geoffroy Oudoumanessah, Carole Lartizien, and 2 more authors2025A key challenge in maximizing the benefits of Magnetic Resonance Imaging (MRI) in clinical settings is to accelerate acquisition times without significantly degrading image quality. This objective requires a balance between under-sampling the raw k-space measurements for faster acquisitions and gathering sufficient raw information for high-fidelity image reconstruction and analysis tasks. To achieve this balance, we propose to use sequential Bayesian experimental design (BED) to provide an adaptive and task-dependent selection of the most informative measurements. Measurements are sequentially augmented with new samples selected to maximize information gain on a posterior distribution over target images. Selection is performed via a gradient-based optimization of a design parameter that defines a subsampling pattern. In this work, we introduce a new active BED procedure that leverages diffusion-based generative models to handle the high dimensionality of the images and employs stochastic optimization to select among a variety of patterns while meeting the acquisition process constraints and budget. So doing, we show how our setting can optimize, not only standard image reconstruction, but also any associated image analysis task. The versatility and performance of our approach are demonstrated on several MRI acquisitions.
@misc{iollo2025activemriacquisitiondiffusion, title = {Active MRI Acquisition with Diffusion Guided Bayesian Experimental Design}, author = {Iollo, Jacopo and Oudoumanessah, Geoffroy and Lartizien, Carole and Dojat, Michel and Forbes, Florence}, year = {2025}, eprint = {2506.16237}, archiveprefix = {arXiv}, primaryclass = {cs.LG}, category = {in_progress}, }
Publications ✅
2026
-
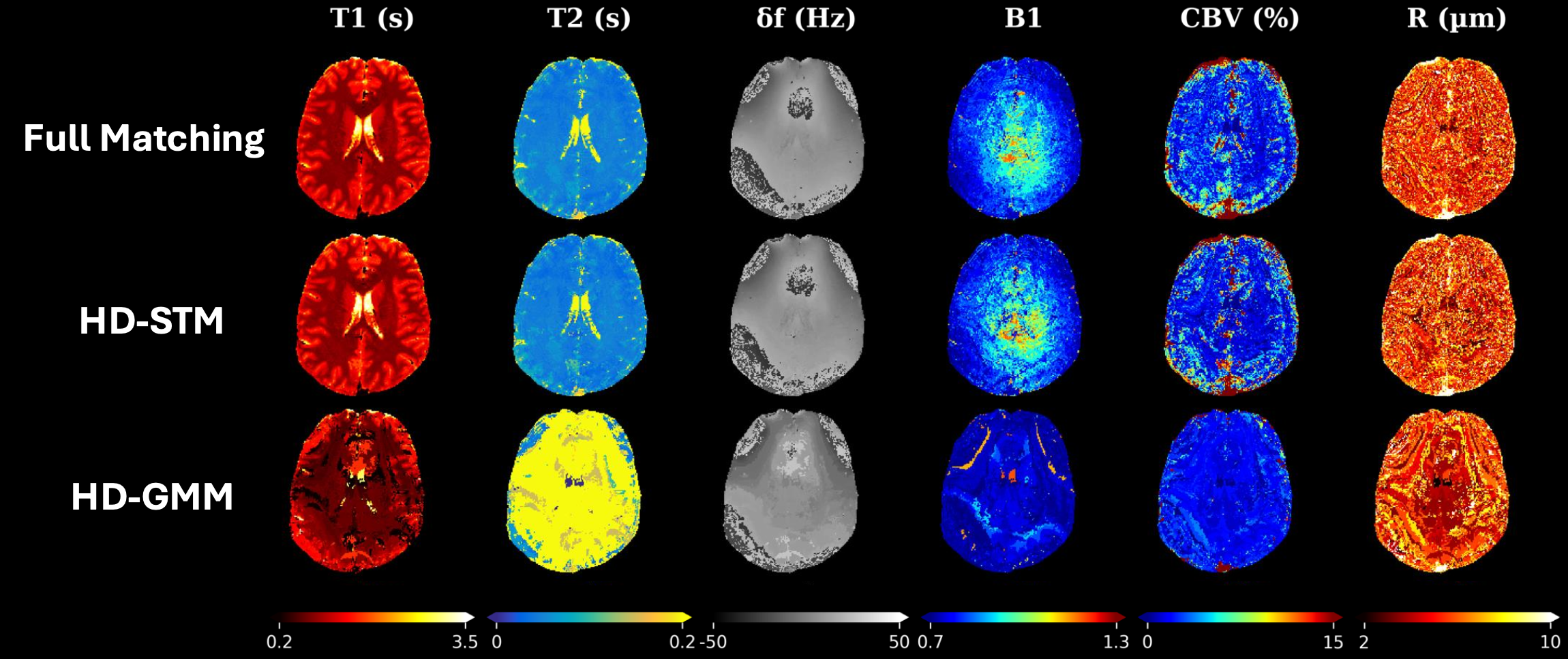 Scalable magnetic resonance fingerprinting: Incremental inference of high dimensional elliptical mixtures from large data volumesGeoffroy Oudoumanessah, Thomas Coudert, Carole Lartizien, and 3 more authorsAnnals of Applied Statistics, 2026
Scalable magnetic resonance fingerprinting: Incremental inference of high dimensional elliptical mixtures from large data volumesGeoffroy Oudoumanessah, Thomas Coudert, Carole Lartizien, and 3 more authorsAnnals of Applied Statistics, 2026Magnetic Resonance Fingerprinting (MRF) is an emerging technology with the potential to revolutionize radiology and medical diagnostics. In comparison to traditional magnetic resonance imaging (MRI), MRF enables the rapid, simultaneous, non-invasive acquisition and reconstruction of multiple tissue parameters, paving the way for novel diagnostic techniques. In the original matching approach, reconstruction is based on the search for the best matches between in vivo acquired signals and a dictionary of high-dimensional simulated signals (fingerprints) with known tissue properties. A critical and limiting challenge is that the size of the simulated dictionary increases exponentially with the number of parameters, leading to an extremely costly subsequent matching. In this work, we propose to address this scalability issue by considering probabilistic mixtures of high-dimensional elliptical distributions, to learn more efficient dictionary representations. Mixture components are modelled as flexible ellipitic shapes in low dimensional subspaces. They are exploited to cluster similar signals and reduce their dimension locally cluster-wise to limit information loss. To estimate such a mixture model, we provide a new incremental algorithm capable of handling large numbers of signals, allowing us to go far beyond the hardware limitations encountered by standard implementations. We demonstrate, on simulated and real data, that our method effectively manages large volumes of MRF data with maintained accuracy. It offers a more efficient solution for accurate tissue characterization and significantly reduces the computational burden, making the clinical application of MRF more practical and accessible.
@article{oudoumanessah2024scalable, title = {Scalable magnetic resonance fingerprinting: Incremental inference of high dimensional elliptical mixtures from large data volumes}, author = {Oudoumanessah, Geoffroy and Coudert, Thomas and Lartizien, Carole and Dojat, Michel and Christen, Thomas and Forbes, Florence}, journal = {Annals of Applied Statistics}, year = {2026}, category = {publication}, }
2025
-
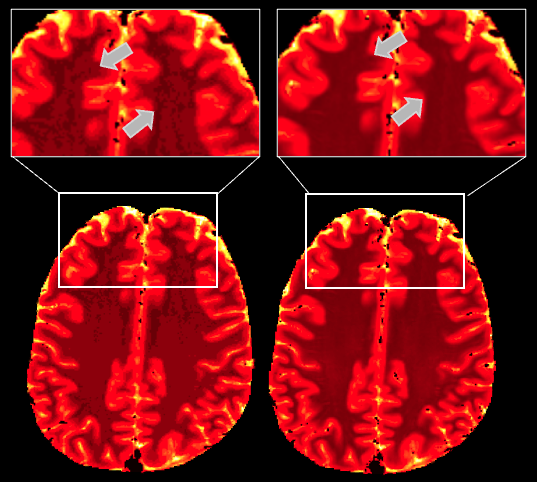 Cluster globally, Reduce locally: Scalable efficient dictionary compression for magnetic resonance fingerprintingGeoffroy Oudoumanessah, Thomas Coudert, Luc Meyer, and 4 more authorsIn 2025 IEEE 22th International Symposium on Biomedical Imaging (ISBI), 2025
Cluster globally, Reduce locally: Scalable efficient dictionary compression for magnetic resonance fingerprintingGeoffroy Oudoumanessah, Thomas Coudert, Luc Meyer, and 4 more authorsIn 2025 IEEE 22th International Symposium on Biomedical Imaging (ISBI), 2025With the rapid advancements in medical data acquisition and production, increasingly richer representations exist to characterize medical information. However, such large-scale data do not usually meet computing resource constraints or algorithmic complexity, and can only be processed after compression or reduction, at the potential loss of information. In this work, we consider specific Gaussian mixture models (HD-GMM), tailored to deal with high dimensional data and to limit information loss by providing component-specific lower dimensional representations. We also design an incremental algorithm to compute such representations for large data sets, overcoming hardware limitations of standard methods. Our procedure is illustrated in a magnetic resonance fingerprinting study, where it achieves a 97% dictionary compression for faster and more accurate map reconstructions.
@inproceedings{oudoumanessah2024cluster, title = {Cluster globally, Reduce locally: Scalable efficient dictionary compression for magnetic resonance fingerprinting}, author = {Oudoumanessah, Geoffroy and Coudert, Thomas and Meyer, Luc and Delphin, Aurelien and Dojat, Michel and Lartizien, Carole and Forbes, Florence}, booktitle = {2025 IEEE 22th International Symposium on Biomedical Imaging (ISBI)}, year = {2025}, organization = {IEEE}, category = {publication}, }
2024
-
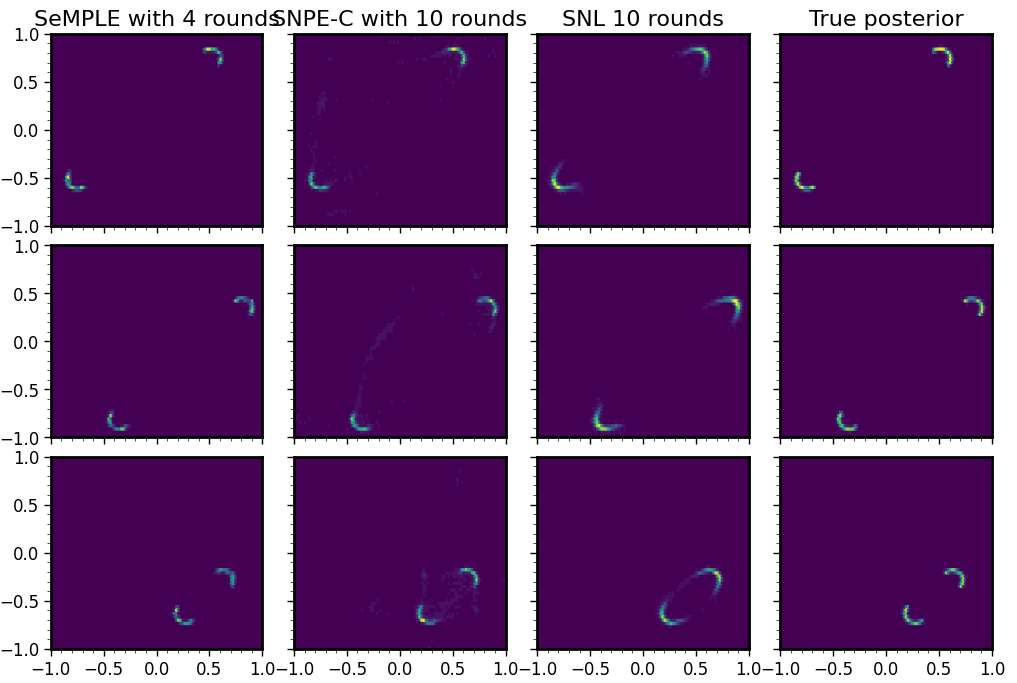 Fast, accurate and lightweight sequential simulation-based inference using Gaussian locally linear mappingsHenrik Häggström, Pedro L. C. Rodrigues, Geoffroy Oudoumanessah, and 2 more authorsTransactions on Machine Learning Research, 2024
Fast, accurate and lightweight sequential simulation-based inference using Gaussian locally linear mappingsHenrik Häggström, Pedro L. C. Rodrigues, Geoffroy Oudoumanessah, and 2 more authorsTransactions on Machine Learning Research, 2024Bayesian inference for complex models with an intractable likelihood can be tackled using algorithms performing many calls to computer simulators. These approaches are collectively known as simulation-based inference (SBI). Recent SBI methods have made use of neural networks (NN) to provide approximate, yet expressive constructs for the unavailable likelihood function and the posterior distribution. However, they do not generally achieve an optimal trade-off between accuracy and computational demand. In this work, we propose an alternative that provides both approximations to the likelihood and the posterior distribution, using structured mixtures of probability distributions. Our approach produces accurate posterior inference when compared to state-of-the-art NN-based SBI methods, while exhibiting a much smaller computational footprint. We illustrate our results on several benchmark models from the SBI literature.
@article{haggstrom2024fast, title = {Fast, accurate and lightweight sequential simulation-based inference using Gaussian locally linear mappings}, author = {H{\"a}ggstr{\"o}m, Henrik and Rodrigues, Pedro L. C. and Oudoumanessah, Geoffroy and Forbes, Florence and Picchini, Umberto}, journal = {Transactions on Machine Learning Research}, issn = {2835-8856}, year = {2024}, url = {https://openreview.net/forum?id=Q0nzpRcwWn}, category = {publication}, }
2023
-
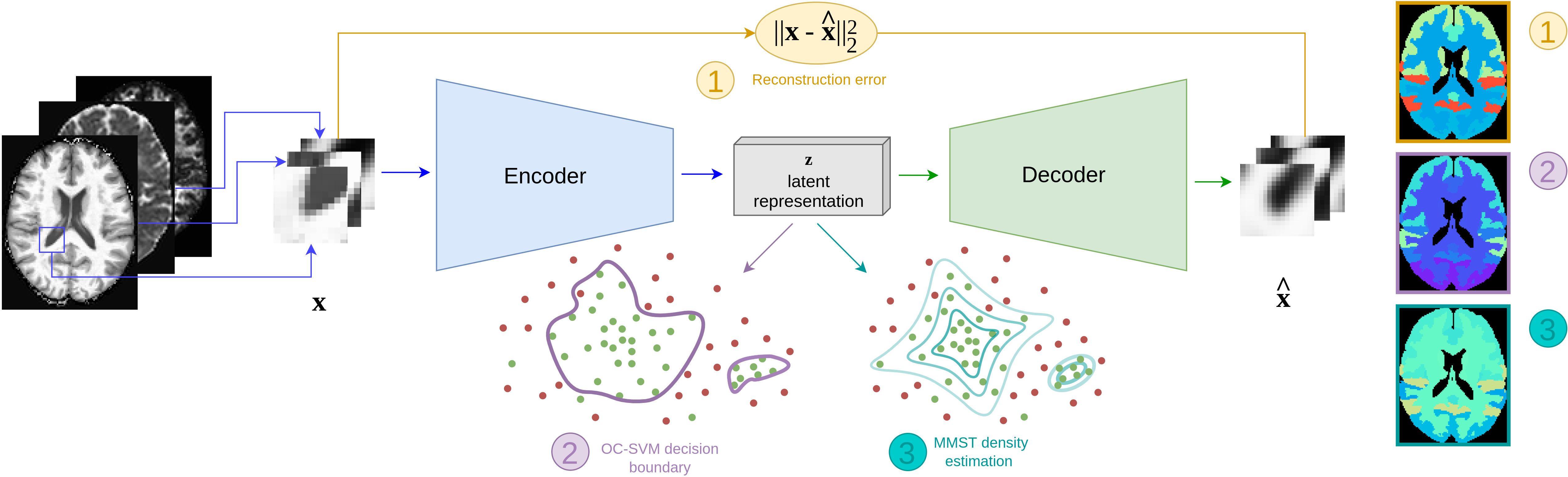 Brain subtle anomaly detection based on auto-encoders latent space analysis: application to de novo parkinson patientsNicolas Pinon, Geoffroy Oudoumanessah, Robin Trombetta, and 3 more authorsIn 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), 2023
Brain subtle anomaly detection based on auto-encoders latent space analysis: application to de novo parkinson patientsNicolas Pinon, Geoffroy Oudoumanessah, Robin Trombetta, and 3 more authorsIn 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), 2023Neural network-based anomaly detection remains challenging in clinical applications with little or no supervised information and subtle anomalies such as hardly visible brain lesions. Among unsupervised methods, patch-based auto-encoders with their efficient representation power provided by their latent space, have shown good results for visible lesion detection. However, the commonly used reconstruction error criterion may limit their performance when facing less obvious lesions. In this work, we design two alternative detection criteria. They are derived from multivariate analysis and can more directly capture information from latent space representations. Their performance compares favorably with two additional supervised learning methods, on a difficult \textitde novo Parkinson Disease (PD) classification task.
@inproceedings{pinon2023brain, title = {Brain subtle anomaly detection based on auto-encoders latent space analysis: application to de novo parkinson patients}, author = {Pinon, Nicolas and Oudoumanessah, Geoffroy and Trombetta, Robin and Dojat, Michel and Forbes, Florence and Lartizien, Carole}, booktitle = {2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI)}, pages = {1--5}, year = {2023}, organization = {IEEE}, category = {publication}, } -
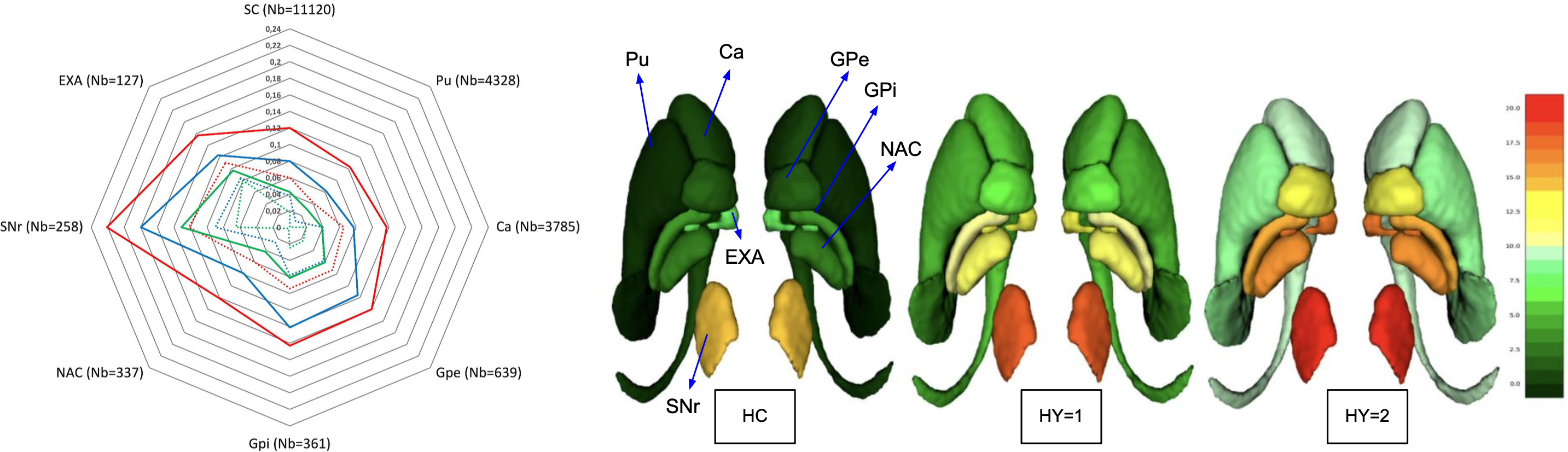 Towards frugal unsupervised detection of subtle abnormalities in medical imagingGeoffroy Oudoumanessah, Carole Lartizien, Michel Dojat, and 1 more authorIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2023
Towards frugal unsupervised detection of subtle abnormalities in medical imagingGeoffroy Oudoumanessah, Carole Lartizien, Michel Dojat, and 1 more authorIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2023Anomaly detection in medical imaging is a challenging task in contexts where abnormalities are not annotated. This problem can be addressed through unsupervised anomaly detection (UAD) methods, which identify features that do not match with a reference model of normal profiles. Artificial neural networks have been extensively used for UAD but they do not generally achieve an optimal trade-off between accuracy and computational demand. As an alternative, we investigate mixtures of probability distributions whose versatility has been widely recognized for a variety of data and tasks, while not requiring excessive design effort or tuning. Their expressivity makes them good candidates to account for complex multivariate reference models. Their much smaller number of parameters makes them more amenable to interpretation and efficient learning. However, standard estimation procedures, such as the Expectation-Maximization algorithm, do not scale well to large data volumes as they require high memory usage. To address this issue, we propose to incrementally compute inferential quantities. This online approach is illustrated on the challenging detection of subtle abnormalities in MR brain scans for the follow-up of newly diagnosed Parkinsonian patients. The identified structural abnormalities are consistent with the disease progression, as accounted by the Hoehn and Yahr scale.
@inproceedings{oudoumanessah2023towards, title = {Towards frugal unsupervised detection of subtle abnormalities in medical imaging}, author = {Oudoumanessah, Geoffroy and Lartizien, Carole and Dojat, Michel and Forbes, Florence}, booktitle = {International Conference on Medical Image Computing and Computer-Assisted Intervention}, pages = {411--421}, year = {2023}, organization = {Springer}, category = {publication}, } -
 Estimation incrémentale pour la détection non supervisée d’anomalies multivariées en imagerie médicaleGeoffroy Oudoumanessah, Carole Lartizien, Michel Dojat, and 1 more authorIn GRETSI 2023-XXIXème Colloque Francophone de Traitement du Signal et des Images, 2023
Estimation incrémentale pour la détection non supervisée d’anomalies multivariées en imagerie médicaleGeoffroy Oudoumanessah, Carole Lartizien, Michel Dojat, and 1 more authorIn GRETSI 2023-XXIXème Colloque Francophone de Traitement du Signal et des Images, 2023Unsupervised anomaly detection in medical imaging is a complex task. State of the art models based on artificial neural networks obtain reasonable performances but at the cost of a high consumption of computational resources. We show that statistical mixture models of probability distributions, in particular multivariate Gaussian mixtures or generalized Student’s distributions, are equally efficient, frugal alternatives without excessive parameterization. We propose an original incremental approach that sequentially estimates the hyperparameters and thus allows to handle large volumes of data. We illustrate the efficiency of this approach with a problem of detecting subtle anomalies in brain MRIs of newly diagnosed Parkinson’s patients.
@inproceedings{oudoumanessah2023estimation, title = {Estimation incr{\'e}mentale pour la d{\'e}tection non supervis{\'e}e d'anomalies multivari{\'e}es en imagerie m{\'e}dicale}, author = {Oudoumanessah, Geoffroy and Lartizien, Carole and Dojat, Michel and Forbes, Florence}, booktitle = {GRETSI 2023-XXIX{\`e}me Colloque Francophone de Traitement du Signal et des Images}, pages = {1--4}, year = {2023}, category = {publication}, }
Abstract ⏱️
2025
-
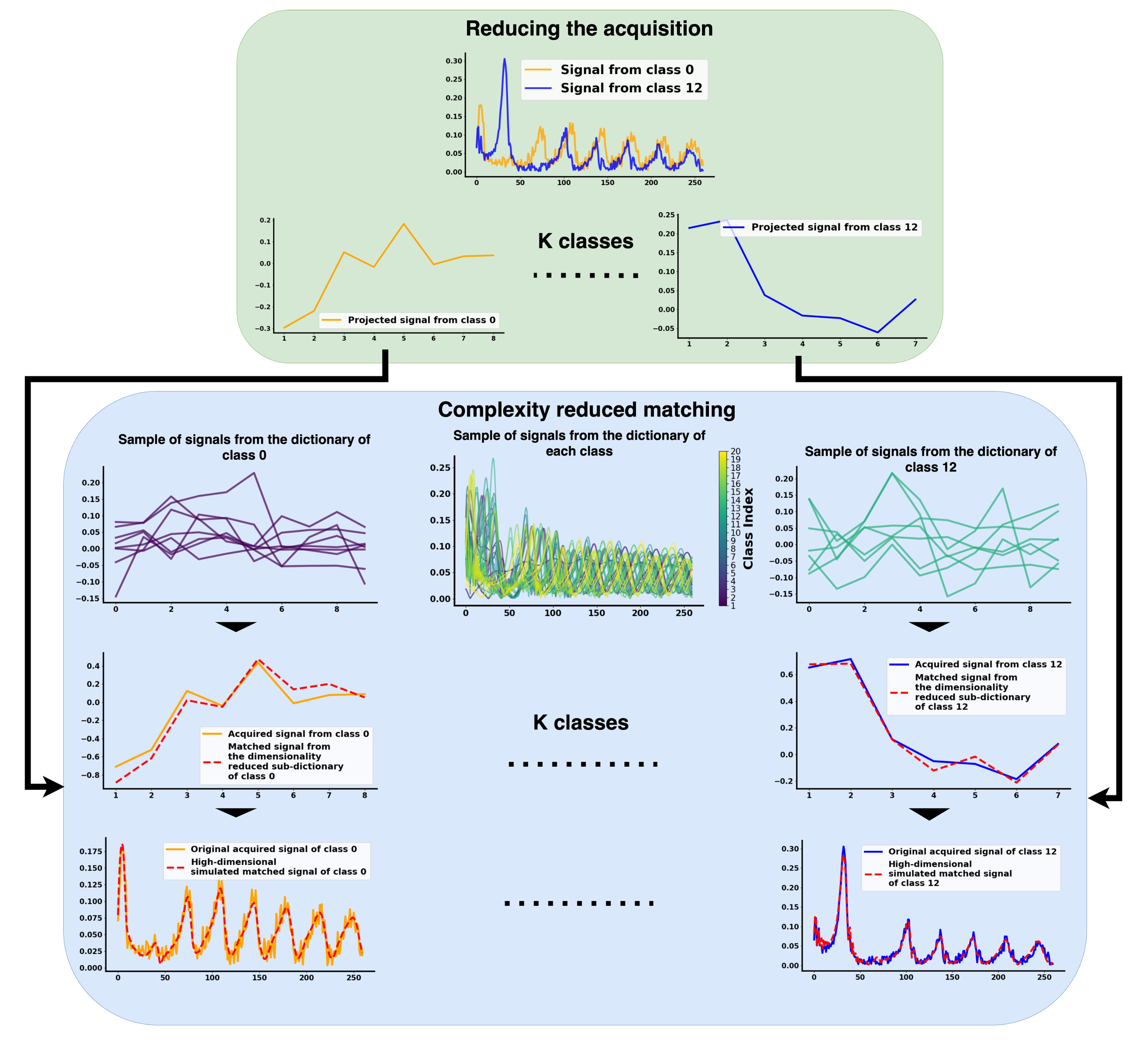 Robust Subspace Clustering Approach for High-Dimensional MRF: Novel Simultaneous Clustering and Dimensionality Reduction at ScaleGeoffroy Oudoumanessah, Thomas Coudert, Antoine Barrier, and 6 more authors2025 meeting of the International Society for Magnetic Resonance in Medicine, 2025
Robust Subspace Clustering Approach for High-Dimensional MRF: Novel Simultaneous Clustering and Dimensionality Reduction at ScaleGeoffroy Oudoumanessah, Thomas Coudert, Antoine Barrier, and 6 more authors2025 meeting of the International Society for Magnetic Resonance in Medicine, 2025@article{oudoumanessah2024robust, title = {Robust Subspace Clustering Approach for High-Dimensional MRF: Novel Simultaneous Clustering and Dimensionality Reduction at Scale}, author = {Oudoumanessah, Geoffroy and Coudert, Thomas and Barrier, Antoine and Delphin, Aurelien and Lartizien, Carole and Dojat, Michel and L. Barbier, Emmanuel and Christen, Thomas and Forbes, Florence}, journal = {2025 meeting of the International Society for Magnetic Resonance in Medicine}, year = {2025}, category = {abstract}, }
Report 🎓
2022
-
 Unsupervised scalable anomaly detection: application to medical imagingGeoffroy Oudoumanessah, Michel Dojat, and Florence Forbes2022
Unsupervised scalable anomaly detection: application to medical imagingGeoffroy Oudoumanessah, Michel Dojat, and Florence Forbes2022@techreport{oudoumanessah2022unsupervised, title = {Unsupervised scalable anomaly detection: application to medical imaging}, author = {Oudoumanessah, Geoffroy and Dojat, Michel and Forbes, Florence}, year = {2022}, school = {Inria Grenoble-Rh{\^o}ne-Alpes; Grenoble Institut des neurosciences}, category = {report}, }